- Research
- Open access
- Published:
Executing cyclic scientific workflows in the cloud
Journal of Cloud Computing volume 10, Article number: 25 (2021)
Abstract
We present an algorithm and a software architecture for a cloud-based system that executes cyclic scientific workflows whose structure may change during run time. Existing approaches either rely on workflow definitions based on directed acyclic graphs (DAGs) or require workarounds to implement cyclic structures. In contrast, our system supports cycles natively, avoids workarounds, and as such reduces the complexity of workflow modelling and maintenance. Our algorithm traverses workflow graphs and transforms them iteratively into linear sequences of executable actions. We call these sequences process chains. Our software architecture distributes the process chains to multiple compute nodes in the cloud and oversees their execution. We evaluate our approach by applying it to two practical use cases from the domains of astronomy and engineering. We also compare it with two existing workflow management systems. The evaluation demonstrates that our algorithm is able to execute dynamically changing workflows with cycles and that design and maintenance of complex workflows is easier than with existing solutions. It also shows that our software architecture can run process chains on multiple compute nodes in parallel to significantly speed up the workflow execution. An implementation of our algorithm and the software architecture is available with the Steep Workflow Management System that we released under an open-source license. The resources for the first practical use case are also available as open source for reproduction.
Introduction
Task automation has a long history in computer science. With the continuing growth in global data, the need for automated data processing becomes more and more evident. This applies to areas such as Bioinformatics [1], Geology [2], and Geoinformatics [3, 4] but also Astronomy [5] (see also “Use case 1: computing astronomical image mosaics” section) and Engineering (“Use case 2: shape optimisation via structural analysis” section). Applications in these areas often employ a number of processing steps (hereafter referred to as actions) that need to be run in a specific order to transform a set of input data into a desired output. A scientific workflow is a model that describes such a transformation. It is typically defined by a scientist using a directed acyclic graph (DAG). This graph specifies the dependencies between the individual actions and how the data flows from one to another in order to produce the desired outcome.
Processing big data sets with complex workflows often requires an amount of resources (in terms of CPU power, available main memory, or hard drive space) that exceeds the capacities of local workstations. Distributed scientific workflow management systems (e.g. Pegasus [6] and Apache Airflow [7]) as well as other solutions for big data processing such as Apache Spark [8] and Apache Flink [9] therefore try to leverage the power of high-performance computing (HPC) clusters and grids, or even more dynamic environments such as the cloud. They traverse the workflow definition and execute individual actions in parallel on distributed compute nodes. This enables scalability [10, 11] and elasticity [12], which allow complex workflows to process large amounts of data in a reasonable time.
Many existing solutions require the workflow designer to model a DAG in its entirety including all actions as well as their dependencies in advance. However, there are use cases that require a more dynamic approach where the number of instances of a given action is unknown at workflow design time or even during run time—for example, if the results of an action should be processed in parallel by a subsequent one but the number of results is not known until the action has actually been executed. In this case, the structure of the workflow has to be adjusted during run time. There are even use cases that involve cycles, iterations, or recursion in the workflow, which cannot be fully modelled with a directed acyclic graph in advance.
Although this issue can be solved in some existing solutions through workarounds or special language constructs, the underlying data models and programming paradigms are often complex and hard to learn and maintain. In summary, this makes design and maintenance of complex dynamic workflows unnecessarily lengthy and tedious (as we show in “Comparison with Pegasus” and “Comparison with Argo” sections).
Goals and contributions
Figure 1a shows a DAG that describes a simple workflow. The input data is first processed by action A. This action produces exactly two outputs, which are in turn processed by two instances of action B. The outputs of these instances are then joined by action C to produce the final result.
Examples of workflow graphs. Petri Net notation according to van der Aalst & van Hee [14] (see also “Workflow model” section)
Russell et al. have identified a pattern for workflows like this, which they call “Multiple instances with a priori design-time knowledge” [13] meaning that the structure of the workflow can be fully modelled before it is executed. A more complex pattern is “Multiple instances with a priori run-time knowledge” shown in Fig. 1b. In this workflow, the number of outputs produced by action A is variable and the number of instances of action B is not known at design time. One reason for this might be that the number of outputs depends on the input—for example, if you want to split a file line by line but do not know how many lines it contains in advance. The number of instances of action B is only known after the execution of action A is completed.
The problem becomes even more complex if the workflow structure changes during its execution. This might be the case if the workflow graph is not acyclic (i.e. if it is not a DAG) but contains loops or recursion. Russell et al. call this “Multiple instances without a priori run-time knowledge”. Figure 1c shows an example, where the data flow is determined by the output of action C. The workflow either finishes or the output of C is passed back to A, in which case the workflow starts again at the beginning. The number of instances of B but also of A and C depends on how many times the workflow repeats. Note that the number of repetitions and thus the final structure of the workflow is unknown, even during run time, until C decides to finish the processing.
As stated above, many existing solutions require the workflow designers to use workarounds to implement Multiple instances without a priori run-time knowledge, if it is supported at all (as shown in “Related work”, “Comparison with Pegasus”, and “Comparison with Argo” sections). It would be beneficial for the designers if the workflow management system would support cycles in workflow graphs and handle Multiple instances without a priori run-time knowledge natively. This would make workflow definitions shorter, so that the designers could focus on the actual task, i.e. the workflow design, and not on technical implementation details.
The two research questions of this paper are therefore as follows:
1) Is it possible to create a distributed scientific workflow management system that natively supports cyclic scientific workflows involving multiple instances of workflow actions without a priori run-time knowledge?
2) Can this system be designed in a way so that the native support for cyclic workflows makes workflow definition and maintenance less complex than with existing solutions?
To answer these questions, we first present an algorithm that splits workflow graphs into smaller process chains suitable for being executed in parallel in a distributed environment. We also present a software architecture for a scientific workflow management system and show how it can be deployed to the cloud. The system uses a workflow model that includes recursive for-each actions. These actions can be dynamically unfolded by our algorithm during run time to support an arbitrary number of iterations.
We also present results from evaluating our algorithm and software architecture by implementing two real-world use cases from Astronomy and Engineering. We focus on the cloud but our approach can also be applied to clusters or grids.
Finally, we compare our system with two existing ones to demonstrate that our approach actually supports cyclic workflows natively and that it does not require workarounds and therefore reduces the maintenance efforts for workflow designers.
An implementation of our algorithm and software architecture is available with the Steep Workflow Management System, which we have released under an open-source license [15].
Structure of the paper
The remainder of this paper is structured as follows. First, we discuss related work and specifically compare our approach with existing scientific workflow management systems (“Related work” section). We then introduce our workflow model and the graphical notation we use to specify workflow graphs (“Workflow model” section).
In the main part of the paper, we present our algorithm to transform cyclic workflow graphs into executable units called process chains (“Workflow algorithm” section). We also describe the individual components of our software architecture and how they work together to execute cyclic workflows in a distributed environment (“Software architecture” section).
After this, we present the results from evaluating our approach and our open-source implementation by applying it to two practical use cases from the domains of astronomy and engineering and by comparing it to two existing systems (“Evaluation” section). We finish the paper with conclusions and future research opportunities (“Conclusions” section).
Related work
The topic of scientific workflow management is of constant and ongoing interest in the research community. There are many open challenges and research questions [16] that have led to a large number of existing implementations. In this section, we focus on the most popular ones, although there are many others still noteworthy such as Galaxy [17], Luigi [18], or Hyperflow [19]. Also, we only discuss the aspect of cycles resulting in “Multiple instances without a priori run-time knowledge” and do not compare other features of the existing implementations.
The main goal of our paper is to support cyclic workflows in a distributed environment natively in order to make workflow definitions shorter and easier to maintain. Existing solutions require workarounds, have limitations, or are not workflow management systems. This section describes the characteristics of each system. The differences are summarised in Table 1.
The first workflow management system that requires workarounds for cyclic workflows is Pegasus [6]. It is well known for executions in distributed environments and has been used in a wide range of applications. However, Pegasus requires the users to specify the entire workflow with a priori design-time knowledge. The system does not have support for a dynamic number of iterations as we propose it in this paper. Instead, users need to apply a workaround and generate a sub-workflow whose structure depends on the results of an action in the parent workflow. For example, if they wish to apply an action to all lines in a file, they first have to create a parent workflow that counts the lines. Then, they need to plan a new DAG that executes the action as many times as there are lines. This approach enables workflows with “Multiple instances with a priori run-time knowledge”. “Multiple instances without a priori run-time knowledge” are also possible by generating a sub-workflow that in turn generates sub-sub-workflows and so on. In contrast to our approach, this workaround is rather tedious to implement and makes maintenance of large workflows hard (see also “Comparison with Pegasus” and “Comparison with Argo” sections).
The second workflow management system that requires workarounds is Apache Airflow [7]. It is has a powerful scheduling interface to support recurring tasks and allows running jobs to be monitored in a web front-end. Similar to Pegasus, workflow modelling in Airflow is based on a DAG. This restricts Airflow in the same way as discussed for Pegasus. The dynamic changes in the workflow structure resulting from cycles can not be realised natively, but require the same workaround using sub-workflows.
Swift/T [20] is more likely to be considered a programming language than a workflow management system. Besides Swift/T, there is also Swift/K [21], which uses Karajan [22] as its underlying workflow engine. Compared to Pegasus and Airflow, Swift/T evaluates the workflow during run time. It is not modelled as a DAG in advance. This enables it to support “Multiple instances with a priori run-time knowledge” natively. “Multiple instances without a priori run-time knowledge” can be designed with limitations: Swift/T has for loops that allow the number of iterations to be modified during run time but do not support distributed execution. For this, Swift/T provides the foreach statement. Nesting foreach inside for combines the benefits of both constructs, but the degree of parallelism is limited. Before the next iteration of the outer for loop can start, all inner iterations of the foreach have to be completed. The for-each actions in our approach support both dynamic number of iterations and parallel execution.
Chiron [23] and the cloud middleware SciCumulus [24] support “Multiple instances without a priori run-time knowledge”. The system uses a data-centric approach [25] for processing. All information are stored in a workflow database and thus can be changed during run time. In [26], this feature is used to reduce the data while it is already processed. However, instead of removing, data can also be added. This leads to new instances without a priori run-time knowledge. If needed, execution can be adapted through so called knops [27] and dynamic loops [28]. Knops enable the user to change parameters during run time. For example, this is required to manually stop an optimization program when the values are good enough. Dynamic loops are even more powerful. They allow the user to interact with the workflow and change the data and control flow during run time. However, the version of Chiron with dynamic loops was never released. It exists as an unpublished development version, but is not available for end usersFootnote 1. Our approach is production-ready and published on GitHub [15].
Argo is a workflow management system based on Kubernetes. The workflow is defined as a Kubernetes Custom Resource Definition (CRD) and uses containers for the actions. If one action outputs a JSON array, Argo can iterate over its entries. This enables “Multiple instances with a priori run-time knowledge”. “Multiple instances without a priori run-time knowledge” can be realised when combining loops with recursion. In Argo, an action can recursively call another action based on defined conditions. This can be used to feed back the results of an action into a loop. However, Argo overwrites the intermediate results of one iteration when the next iteration starts. The developer has to introduce and update a custom counter variable to write the intermediate results to different files. Additionally, the termination of the recursion can not be decided based on the existence of results. The developer has to write a custom action to check if there are results or use a read error as a termination condition. For more information, see “Comparison with Argo” section.
Taverna [29] uses a GUI-based modelling approach. It has a strong focus on reusability of components and is widely used in bioinformatics. Like Pegasus and Airflow, a workflow is modelled as a DAG. However, Taverna extends this model by loops. The developer is able to define a sub-workflow where the input parameters are a subset of the output parameters. A boolean expression defines whether the current output values should be used as input values for a new iteration or the loop ends. Parallelism is realized by using a list of input values for an action that requires a single value. In this way, “Multiple instances without a priori run-time knowledge” can be modelled more easily because sub-sub-workflows are no longer needed. However, Taverna has a drawback: it is not a distributed system. It runs on a single machine and can only connect to external WSDL (Web Services Description Language) services. Distribution requires third-party components such as Tavaxy [30].
Kepler [31] has a user interface that allows users with no or little background in computer science to build their own workflow. A workflow in Kepler consists of actors. An actor can be a script or an expression to decide whether a loop should be finished or not. This enables “Multiple instances without a priori run-time knowledge”. The execution of the actors is controlled by directors. Kepler has many integrated directors for different use cases. For a parallel execution, the PN director can be used. It encapsulates each actor in its own thread, enabling several actors to run simultaneously without blocking each other. Compared to other workflow management systems, distributed execution is not as easy to achieve. Like Taverna, Kepler can execute WSDL services and access data in a grid. Additionally, services can be invoked via REST calls, Soaplab or Opal. However, it is not possible to distribute actors to multiple cloud machines directly. Nevertheless, there are approaches to use Amazon EC2 instances and access them via SSH [32].
Besides these frameworks, there are programming models like MapReduce [33] and corresponding implementations such as Hadoop [34] and Apache Spark [8]. Wang et al. have combined Kepler with Hadoop to allow it to make use of distributed computing [35]. Fei et al. presented an approach to combine the strengths of MapReduce with a scientific workflow management system [36].
Especially for long running scientific tasks, Spark scales well [37] but requires a workflow definition using its own API. This problem was solved by Gaspar et al. who presented an approach to run existing workflows inside a Spark cluster [38]. They are able to use the optimisations in Spark regarding data locality, while their workflows do not need to be rewritten. However, scientists with no knowledge in distributed computing may be unable to use these alternative programming models correctly. A workflow management system can provide benefits to them in terms of usability, scalability, and flexibility [3]. In this paper, we present an algorithm and a software architecture for such a scientific workflow management system.
Workflow model
This section gives a brief overview of the workflow model in our system and the notation we use to present workflow graphs in this paper. Compared to existing workflow definition languages such as YAWL [39] or CWL [40], our workflow model is lightweight. CWL also does not support loops or recursion [41]. Our model resembles the workflow description language WDL [42] but inputs and outputs are not strongly typed. Also, instead of scatter or gather blocks to express parallelisation and joins, there are only two, very generic types of actions.
Figure 2 shows the UML class diagram. A workflow has a human-readable name, a list of variables, and a list of actions. Each variable is a key-value pair, whereas the key is an identifier and the value can be a primitive (boolean, number, string), a list of primitives, or undefined if it is unknown at design time. Variables with undefined values are typically used to create links between the outputs and inputs of actions. Initialized variables are immutable.
The two types of actions in our model are execute actions and for-each actions. Execute actions refer to an externally defined processing service that should run in the cloud. They specify inputs and outputs as well as generic parameters for the service. For-each actions have sub-actions that should be applied to a list of input data. Results are collected in an output list. Each for-each action has an enumerator, which is a variable the sub-actions can use to refer to an item in the input list. The property yieldToOutput points to an output of a sub-action that should be collected in the output list of the for-each action. yieldToInput, on the other hand, allows the workflow designer to implement recursion by feeding the output of a sub-action back into the input list of the for-each action.
The listing shown in Fig. 3 shows a simple example workflow in YAML notation. It consists of a for-each action that iterates over all files in an input directory data_directory. For each file, the service process_file is called. When the loop is complete, all outputs are processed by the service aggregate and a final_result file is created.
More information about the data model including examples can be found in the documentation of our open-source implementation [15].
In order to graphically express scientific workflows, we use Petri Nets [43–46] with the extensions by van der Aalst & van Hee who define special symbols for common constructions [14]. The following table lists the main symbols and how we use them in this paper:
-
 AND-split
AND-splitAn action that produces multiple results at the same time. It will write to all defined outputs.
-
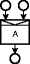 AND-join
AND-joinAn action that has multiple inputs. The action can only be executed if all inputs are available.
-
 OR-split
OR-splitAn action that has several defined outputs but produces exactly one result. The action decides itself to which output it will write the result.
-
 AND/OR-split
AND/OR-splitAn action that will write a result to one or more of its defined outputs at the same time.
In order to express loops and multiple instances more concisely than in the examples in Fig. 1, we specify an additional symbol. It consists of a box with a circular arrow in the upper-left corner. Inside this box is a nested Petri Net with the transitions that should be repeated or, more precisely, parallelised. In our case, the symbol represents a for-each action with the nested Petri Net being the sub-actions.
-
 For-each action
For-each actionA for-each action with a nested Petri Net. The action will apply the sub-actions to all input values (typically in parallel). The results will be collected in the output of the for-each action.
Figure 4 shows some examples. With the new symbol, we can represent all elements of our workflow model: a place (circle) corresponds to a variable, a transition (rectangle) is an execute action, and the new symbol represents a for-each action. Recursion with yieldToInput can be modelled by combining the new symbol with anB OR-split.
Note that, in order to maintain the semantics of Petri Nets, we need to add artificial, no-operation transitions represented by black lines. These transitions do not appear as actions in our workflow. As will become clear in the following sections, they can be seen as synchronization points where our algorithm splits the workflow or where it processes the results of the sub-actions of a for-each action.
Workflow algorithm
The main purpose of our algorithm is to split a (possibly cyclic) workflow graph into smaller pieces, which we call process chains. A process chain is a linear list of actions (including all values of input and output parameters) that should be executed on the cloud nodes.
Our algorithm is meant to be used iteratively: we first call it to generate process chains, then execute them, and finally feed the results back into the algorithm to create more process chains (see “Software architecture” section for details). During each iteration, the algorithm removes the actions it was able to convert to process chains from the workflow. This procedure repeats until all actions in the workflow have been consumed or until the algorithm returns no more process chains.
Our algorithm consists of two functions that are called sequentially (see pseudo-code in Fig. 5): unrollForEachActions and generateProcessChains. For the sake of clarity, we first describe generateProcessChains, which splits a workflow into smaller pieces, and then unrollForEachActions, which implements the main contribution of our paper, namely cyclic workflows.
Generating process chains
Figure 6 shows the pseudo-code of generateProcessChains. The body of the function consists of a for loop that iterates over all execute actions in the workflow. For each of them, it checks if all inputs are available (either because they were statically defined in the workflow or because they have been generated in an earlier iteration of the workflow execution). If they are available, the action is ready to be executed. In the inner while loop, the function then tries to create a linear chain of subsequent actions that are also ready to be executed. An action B is considered subsequent to an action A if all inputs of B are outputs of A. The inner while loop stops if it encounters a junction in the workflow graph (a split or a join) or if it runs out of execute actions.
The function removes actions ready to be executed from the workflow. It also keeps track of actions that it visited but considered not ready to be executed in order to avoid visiting them again. However, the function does not mark an action as visited if it is the first one in a chain because it could still be an action that is subsequent to another one. The function collects all created process chains and returns them in the end.
Example 1
Figure 7 depicts how generateProcessChains splits an example workflow graph into process chains. The input of action A is available from the beginning because it has been statically defined in a workflow variable. Action A is therefore ready to be executed, and the function creates a process chain containing only this action in the first iteration. It then removes A from the workflow. In the second iteration, when the outputs of A have been generated and therefore the inputs of B and D have become available, the function creates two process chains: one containing the consecutive actions B and C, and another one containing D. In the third iteration, the inputs of E have also become available and the algorithm creates the final process chain.
Unrolling for-each actions
As generateProcessChains only handles execute actions, we need a way to convert for-each actions to execute actions. This process is called loop unrolling.
unrollForEachActions (see pseudo-code in Fig. 8) iterates over all for-each actions in the workflow. For this, it uses a queue to be able to add nested for-each actions during the process and to unroll as many levels of nested for-each actions as possible in one call.
The function first checks if all inputs of a given for-each action are availableFootnote 2. If this is the case, the action is ready to be unrolled. Unrolling works as follows: For each input, the function clones all sub-actions and injects the input into the cloned actions. Injecting means that it reassigns the input variables, which point to the enumerator of the for-each action, to the current input. The function then adds the cloned actions to the workflow. Cloned for-each actions are appended to the queue so the function can unroll them too.
The function also iterates over all cloned actions in the workflow and collects all outputs that need to be fed back into the input of the for-each action via yieldToInput in a set of recursive inputs. There are three cases when this set can be empty: the property yieldToInput was not set, all cloned actions have been executed, or all inputs of the for-each action have been consumed. In either case, the for-each action can be removed from the workflow. Otherwise, the function needs to keep it so unrolling can continue in the next iteration of the algorithm.
Example 2: ordinary for-each action
Figure 9 shows an example of how the algorithm splits a workflow graph containing a for-each action. In the beginning, action A is the only one that is ready to be executed. In the first iteration, the algorithm therefore creates one process chain containing A. It does not add other actions because the one subsequent to A is a for-each action (as described earlier, the function generateProcessChains only handles execute actions). If we assume that executing this process chain results in three outputs, the function unrollForEachActions then clones the sub-actions of the for-each action three times and adds the clones to the workflow. It also removes the for-each action. generateProcessChains traverses all execute actions in the workflow (including the new ones) and generates three process chains. These process chains can then be executed in the cloud. After that, the outputs of all instances of action B are available, and, in the third iteration, the algorithm can create a new process chain containing action C. Note that C has only one input, namely a list of the outputs created by all instances of B.
Example 3: recursive for-each action
In the previous example, we described how our algorithm splits a workflow graph containing a for-each action without cycles. We now present an example workflow with a recursive for-each action.
Figure 10 shows the original graph and the individual iterations. The workflow consists of a for-each action with exactly one sub-action C that has two outputs. The left output is connected to the output of the for-each action via yieldToOutput. The right output is connected to the input of the for-each action via yieldToInput.
In the first iteration, given that the input of the for-each action is a list of two items, the function unrollForEachActions clones the sub-action two times and distributes the input items. generateProcessChains then creates a new process chain for each clone and removes the clones from the workflow. Note that the for-each action is not removed yet because the new process chains might produce more items to iterate over.
The two process chains are then executed and their results are passed back to the algorithm. Let us assume that action C1 produced an item in its left and C2 in its right output. The output of C1 is collected in the output of the for-each action while the output of C2 is fed back into the input of the for-each action. In the second iteration, the algorithm then clones the sub-action only once and also creates only one process chain (containing action C3). In the third iteration, assuming C3 has produced an item in its left output, the algorithm removes the for-each action because there are no more inputs to process. The workflow is now empty and no process chains are created. The final output of the for-each action is a list of two items: the output produced by C1 after the first iteration and that of C3 after the second iteration.
Note that the for-each action in this example contains only one sub-action. Nevertheless, our algorithm supports both recursive for-each actions with multiple sub-actions as well as nested sub-actions as shown in the example workflow in Fig. 4c (see our evaluation, in particular the use case in “Use case 2: shape optimisation via structural analysis” section). It further supports nested for-each actions with an arbitrary number of levels. For the sake of brevity, we do not include an example for such a complex workflow here. The image would be several pages long. The general process, however, is just a combination of examples 1, 2, and 3. It requires interaction between both unrollForEachActions and generateProcessChains. We leave the example as an exercise to the reader.
Software architecture
In this section, we describe the main components of the software architecture of our open-source workflow management system where we implemented our algorithm. The components are shown in Fig. 11. The Controller manages the workflow execution. It keeps the workflow and any generated process chains in a database. At the beginning of the workflow execution or when process chain results are available, the Controller calls our algorithm to generate new process chains. In parallel, another component called the Scheduler regularly polls the database, assigns any new process chains to compute nodes in the cloud and then oversees their execution. The Scheduler writes process chain results back to the database.
The purpose of the database is twofold. First, it is an asynchronous communication channel between the Controller and the Scheduler. Second, it enables fault tolerance: If the workflow management system crashes, it can resume workflow execution on restart and does not have to repeat the whole workflow. In the worst case, it only needs to run those process chains again that were currently being executed when the system crashed.
In the following, we describe the behaviour of the Controller and the Scheduler during workflow execution in detail.
Controller
Figure 12 shows the control flow inside the Controller while it executes a single workflow. When the execution is started, the component first calls our algorithm to generate an initial set of process chains. If the algorithm actually has created process chains (i.e. if the workflow is valid and not empty), the Controller registers them in the Database. At this point, the Scheduler starts executing the registered process chains (see “Scheduler” section). The Controller starts a periodic job to look for process chain results in the database. In case of an error, it aborts the workflow execution. Otherwise, it calls our algorithm again to generate more process chains. The whole process repeats until the algorithm does not produce more process chains.
Note that our algorithm may return an empty set of process chains even though the workflow still contains actions. This can happen if the workflow is invalid and there are actions for which there are no inputs—which means the actions are never ready to be executed. If this case is detected at the end of a workflow execution, our implementation marks the workflow as ‘failed’ in the database.
Scheduler
The Scheduler is responsible for overseeing the execution of process chains in the cloud. Figure 13 shows its internal control flow. Basically, the Scheduler works in an infinite loop where it iterates over all new and running process chains currently registered in the database. For each process chain, the control flow can take either of the following two paths:
-
If the process chain is new, the Scheduler selects a compute node and then starts the execution of the process chain on this node. If no node is available (e.g. because all nodes are currently busy), the Scheduler just continues with the next process chain.
-
If the process chain is currently registered as ‘running’ in the database, the Scheduler checks the actual status on the compute node. If the process chain has finished, it registers the results in the database and then continues with the next one. At this point, the Controller can pick up the results and run our algorithm again.
Evaluation
As described earlier, we implemented our algorithm in a workflow management system called Steep, which we released under an open-source license [15]. In this section, we evaluate our approach by applying it to two real-world workflows. We also compare it with the workflow management systems Pegasus (“Comparison with Pegasus” section) and Argo (“Comparison with Argo” section) to further differentiate our approach from existing ones.
Our algorithm, the controller, and the scheduler are implemented in Kotlin using Eclipse Vert.x [47], a tool-kit for building reactive applications on the Java Virtual Machine (JVM). With Vert.x, it is possible to develop distributed applications consisting of multiple instances running on different virtual machines in the cloud and communicating with each other through an event bus. Steep supports multiple database back-ends. For this evaluation, we chose MongoDB [48] because it is fast and lightweight and can be easily deployed in the cloud. We stored the input data as well as intermediate and final results of the workflows in the distributed file system Gluster [49].
Figure 14 shows a deployment template for the different environments we ran our workflows in. The environments differed only in the number of compute nodes, which is indicated by the three dots. We put the binaries of our workflow management system into a Docker image and deployed it to the compute nodes. Note that this means that on every compute node, there were a controller and a scheduler. This allowed us to not only scale the processing services but also the workflow management system itself by increasing the number of compute nodes.
The components shared persistent data in MongoDB, which ran on a separate virtual machine. We also deployed an NGINX proxy running on a gateway machine with a public IP address. This allowed us to start workflows and to query progress from our workstation. We mounted the Gluster file system to all compute nodes as well as the gateway. This enabled all processing services to access the same data. It also allowed us to upload input data to the cloud as well as to fetch workflow results through the gateway.
We created the virtual machines, the block devices, and a virtual private network in the Amazon Web Services (AWS) cloud using Terraform [50]. After this, we deployed our workflow management system, MongoDB, and Gluster using Ansible [51]. Terraform and Ansible are automation tools that allowed us to express the infrastructure and the deployment as code and, in consequence, to quickly reset the environment between workflow runs and to easily change the number of compute instances.
Table 2 shows the individual AWS instance types we used for our virtual machines. For the Gluster file system we created block devices with a capacity of 150 GB each and attached them to the compute nodes as secondary volumes. We configured a replication factor of 2, so the total size of the distributed file system was 150 GB ×n/2 with n being the number of compute nodes. Our operating system was Ubuntu 18.04 LTS.
Use case 1: computing astronomical image mosaics
The first workflow we executed is Montage. It is a well-known workflow that is often used in literature to evaluate distributed scientific workflow management systems. Its main purpose is to create a mosaic of images taken by telescopes from professional astronomical surveys such as the Two Micron All Sky Survey (2MASS) [52]. These images need to be pre-processed before they can be merged into a larger one.
Figure 15 shows the workflow for one colour band. The workflow makes use of processing services from the Montage toolkit [53]. It requires a set of input images and a header template. Both can be created with the toolkit (see below). At the beginning of the workflow, the images are reprojected to the scale defined in the header template (processing service mProject). After that, new metadata is generated with mImgtbl. Based on this, mOverlaps tries to identify overlapping image pairs. For each image pair, the workflow calls mDiffFit, which calculates the image difference.
The result is then processed by a service called joinDiffFitResults. Note that this service is not part of the Montage toolkit. It is a custom, very lightweight service we created. Its sole purpose is to generate the required input parameters for the subsequent service mConcatFit. Other Montage workflow definitions from literature do not include such a service, but they also do not include mOverlaps (e.g. [54–56],). Since the result of mOverlaps determines the number of instances of mDiffFit, existing works need to run mOverlaps prior to the actual workflow execution and therefore model the workflow with a priori design-time knowledge. We show that, with our approach, we can move mOverlaps into the workflow and achieve “Multiple instances with a priori run-time knowledge” instead. This allows us to use the same workflow definition for multiple input data sets, while existing works need to create a new DAG for each input data set (e.g. [57]).
mConcatFit merges the results of all instances of mDiffFit into one file. This file is then used by mBgModel to determine a set of corrections to apply to each image. The next service mBackground uses these corrections to remove the background from each of the input images. After this, new metadata is generated with mImgtbl and the individual images are merged to a mosaic with mAdd. Finally, the result is resized with mShrink and converted to a JPEG image with mViewer.
Before we could test our implementation with this workflow, we needed to find a set of input images and create a header template. The Montage toolkit contains a command called mHdr for the header template, and another one called mArchiveExec that can be used to download imagery from different surveys. We opted for 2MASS because the provided images are public domain and cover the whole sky. We focussed on the Orion Nebula and created two data sets (A and B), which are described in Table 3. Note that the second data set overlaps with the first one, but its centre lies north of the Orion Nebula and the covered area is smaller. Also note that 2MASS provides three colour bands J, H, and K with different wavelengths. Since the workflow shown in Fig. 15 only covers one band, we copied the whole definition two more times and merged the three colour bands at the end of the workflow with mViewer, which is able to take three input channels for red, green, and blue.
We then created 4 compute nodes in the AWS cloud and ran the workflow two times, once for each input data set. Since we automated the deployment with Terraform and Ansible, we were able to reset the environment after each workflow run in order to establish similar conditions. In order to be able to validate that there is a speed-up when parallelising the workflow execution, we also ran the two tests with only 1 compute node. The results of all tests are shown in Table 4.
The parallelised runs are about three times faster than the non-parallelised ones, although we have 4 compute nodes instead of 1. This is primarily caused by the overhead of moving the large input data as well as intermediate results between the nodes but also by the fact that processing services such as mBgModel and mAdd alone take the majority of the run time (1 h each) and can only be parallelised on the level of colour bands, which means a maximum of three parallel instances.
Figure 16 shows the final images for the two input data sets A and B. Note that we put the complete source code of the workflow as well as the Terraform and Ansible configuration files for AWS on GitHub as open source for interested readers who want to reproduce our setup and results [58, 59].
Use case 2: shape optimisation via structural analysis
Our second use case focuses on workflows where the number of iterations and the number of parallel executions per iteration are not known when designing and launching the workflow and can change dynamically during the execution (i.e. without any a priori design-time or run-time knowledge). The aim is to perform a shape optimisation of a given 3D object using structural analysis simulations inside a sparse grid, similar to the approach presented by Tamellini et al. [60]. Such optimisations are performed, for example, in the product development process in the area of mechanical engineering. Given an objective function, the optimisation iteratively tries to find an optimal shape of the 3D object for a pre-defined load case. For every iteration, different variants of the initial object are being simulated in parallel. The number of variants—and therefore the number of parallel executions—depends on the variance between the simulation results of the previous iteration. The optimisation terminates after the simulation results have converged and the variance between them is below a certain threshold.
For our evaluation, we used a Catmull-Clark subdivision solid model [61] of a wrench. On that model, we defined three parameters: the outer diameter of the open end of the wrench, the thickness of the middle section relative to the xy plane, and the thickness of the middle section relative to the xz plane. Figure 17 shows three design variants of the wrench models, each corresponding to different values of these parameters. During the optimisation, different sets of parameter values are evaluated using a structural mechanics simulation. Figure 18 shows the simulation setup with a fixation at the right end of the wrench (shown in orange) and an external force being applied to the left end (yellow box and red arrow). Details on the process of creating and simulating parametrised Catmull-Clark subdivision solid models can be found in the work of Altenhofen et al. [62, 63]. The objective function for the optimisation models a trade-off between stability and weight of the wrench, favouring light designs while ensuring the stress and the deformation under load to be below a given threshold.
Figure 19 shows the workflow in our Petri Net notation. It starts with a processing service called createInitialSamples creating a pre-defined number of initial sample points in the parameter space [0,1]3 along a regular grid. Each point represents a set of parameter values corresponding to a concrete design variant of the wrench to be optimised.
The workflow contains an outer for-each action that is used to model multiple iterations. Its input consists of either one element or none. If there is an element, it performs an iteration. Otherwise, the workflow terminates. Inside, there is another for-each action that performs the simulations on the design variants.
createInitialSamples only creates sample points for the first iteration. It writes them into a single file. Inside the outer for-each action, this file is first split into individual files—one for each sample point—by the splitSamples service. For each sample point, runSimulation performs a structural analysis simulation in the inner for-each action. The different simulations run in parallel. Each simulation writes its results to a separate file.
The service evaluateSimResults reads these files and maps the results to a score value using the objective function. The best sample point defines the centre of a new, smaller grid that is sampled in the next iteration of the optimisation process. 23 to 73 sample points are created, depending on the variance between the score values. Finally, evaluateSimResults creates a new file with the sample points that is fed back into the outer for-each action. The workflow finishes when evaluateSimResults does not create a new file with sample points but instead writes the final parameter set to another file specified by its second output.
In our evaluation setup, the optimisation performed 6 iterations, evaluating a total number of 67 sample points. Figure 20 shows the distribution of sample points inside the parameter space. Figure 21 shows the score values and their variance throughout the iterations. The optimised wrench model is 15.6% lighter compared to a design manually created by the engineer. At the same time, the maximum stress is reduced by 10.7% and the maximum deformation is reduced by 62.2%.
Table 5 shows the run time of the optimisation on different numbers of virtual machines in the Amazon cloud. Please note that the speed-up per machine is reduced when using more than 8 machines since the first iteration evaluates 27 sample points, while the remaining five iterations each evaluate 8 sample points according to a reduced variance between the simulation results. Including the pre- and post-processing services, 80 processes are executed in total.
Comparison with Pegasus
In this section, we compare our approach with Pegasus, a popular workflow management systems, which is open-source and represents other systems that consume DAGs as input very well. We applied Pegasus to both of our use cases.
Use case 1
In the official GitHub organisation of Pegasus [64], there is an implementation of a Montage workflow similar to the one described in “Use case 1: computing astronomical image mosaics” section. The repository consists of a Python script that requires you to specify the area of the sky for the image to create as an argument. From this, it generates a suitable DAG for Pegasus.
The script first downloads the raw images. It then needs to perform further calculations. This is because the service mDiffFit must be executed for each overlap in the raw images during workflow execution. However, which raw images actually overlap can only be determined after their projection, which is done in the workflow as well. This is a problem because the execution of mDiffFit has to be modelled in the DAG in advance. As a solution, the program mDAGTbls is used to generate an estimation of where overlaps exist. This estimation is inaccurate and provides a superset of true overlaps. However, the execution of mDiffFit can be planned based on this estimate. Since the estimation is a superset, no data is lost. Nevertheless, more service calls are planned than would actually be necessary for execution.
Our system, on the other hand, supports “Multiple instances without a priori run-time knowledge” and is able to plan the execution of mDiffFit at run time. No estimation is required before the workflow is executed, and only needed service instances are created. We ran the workflow with the parameters specified in the Pegasus Montage repository and the number of mDiffFit calls were reduced from 900 (Pegasus) to 216 (our approach).
In addition, our approach does not need a setup script at all. Since our system can dynamically adapt the workflow structure during run time, we can even perform the download of the raw images within the workflow (see our implementation of the Montage workflow [58]).
This has multiple advantages: First, it avoids additional development effort during workflow design and reduces maintenance overhead later on because it allows users to focus on the workflow itself without requiring them to write a setup script in a general purpose programming language such as Python.
Furthermore, the computation (and even the download) can be completely executed in the cloud. Also, the setup script of Pegasus cannot be parallelised. Our approach therefore saves resources and also allows for maximum parallelisation (our implementation of the Montage workflow even downloads the raw images in parallel—depending on the Internet connection and the amount of data to be retrieved, this can save a lot of time).
Another benefit is that our approach allows the same workflow to be used for multiple input data sets. It has very few parameters: the name of the survey providing the raw images, the celestial object or the location to focus on, and the width and height of the area of interest in degrees. These parameters are specified as variables in our workflow file. Users can change them and then directly submit the workflow without any further changes on the structure.
Use case 2
The missing support of “Multiple instances without a priori run-time knowledge” in Pegasus complicates the design of our second use case. The number of iterations of the outer loop in Fig. 19 is unknown before execution. Only when the simulation results are available, it can be decided if another iteration is necessary. This is similar to the situation in the Montage workflow, where the number of overlaps is calculated during run time. However, it is not possible to estimate a superset here. The only possibility for an execution using a single DAG would be to define a maximum number of iterations. All these iterations would have to be planned regardless of whether they are needed for a concrete data set or not. Also, the optimisation would not finish correctly if too few iterations were planned.
Alternatively, Pegasus offers the concept of sub-workflows. During the execution of the workflow a new DAG can be created and its execution can be triggered. This way, the simulation results can be evaluated and a new DAG can be planned afterwards. The new DAG either contains another iteration or terminates directly.
We implemented the workflow in Pegasus resulting in the rather complex run time model shown in Fig. 22. The workflow starts on the left hand side (1). It first creates the set of initial sample points and then plans a new DAG with a static number of simulations (based on the number of sample points). It then executes the DAG by submitting it to Pegasus. This leads to a new sub-workflow (2) that performs the actual simulations and evaluates their results. Based on this, another DAG is planned and executed, which again leads to a new sub-workflow. This process repeats recursively (3) and more nested sub-workflows are generated and executed. Since it is not possible to know in advance when the workflow will end, even the final iteration (4) contains actions to plan and execute a new DAG. The structure of a workflow cannot change during run time in Pegasus, so these actions have to be executed, even though there is no more work to do. They just generate and submit a final empty workflow (5).
Note that the first workflow (1) has to wait until all recursively embedded workflows have finished. Depending on the number of iterations, this may lead to a large number of workflows running in parallel.
With our approach, on the other hand, all steps happen in only one workflow. If you compare Fig. 22 with Fig. 19, you will notice that our implementation is significantly less complex. Also, our approach saves resources, does not require a workaround with static sub-workflows, and is easier to design and maintain.
Comparison with Argo
Besides Pegasus, we also compared our approach with the scientific workflow management system Argo, which offers features that are very close to our system. Compared to Pegasus, it even offers constructs to specify loops and recursion. However, these constructs are not as powerful as ours. The two use cases therefore require workarounds that make the workflow definitions longer and more complex.
Use case 1
Argo is based on Kubernetes. Workflows are defined as Kubernetes Custom Resource Definitions (CRD) and actions run inside Docker containers. An action can communicate with Argo by writing a JSON array to its standard output. Argo is able to iterate over the array entries and to dynamically create new containers. This enables “Multiple instances with a priori run-time knowledge”.
Compared to Pegasus, this feature makes it slightly easier to define the Montage workflow because the workflow structure does not have to be specified completely in advance. However, the services from the Montage toolkit do not produce JSON arrays. They write their results to files. In order to make them compatible with Argo’s approach, workflow designers are either required to write wrapper services that transform the output of the services to JSON, or they have to create additional Docker containers that need to be executed after each Montage service to convert the list of files produced by this service to a JSON array.
Figure 23 shows such a Docker container. It is based on the python:alpine3.6 base image and contains a small Python program that reads all files from a directory and prints them as a JSON array.
In contrast, our system supports multiple output files natively and therefore does not require such a workaround.
Use case 2
Use case 2 is more complex than use case 1 as it contains cycles whose number of iterations can change dynamically during run time. Compared to our approach, this feature is not directly supported by Argo. The system supports loops, but these loops can only iterate over a JSON array (as described above) and do not have a dynamic break condition. Nevertheless, this feature can still be used to model the inner for-each action of the workflow of use case 2. However, similar to the Montage workflow, this requires to either change the implementation of the splitSamples service so it writes a JSON array to its standard output, or to implement an additional workflow step starting a container to convert the list of files to an array.
The outer for-each action of the workflow has a dynamic number of iterations. It stops when the evaluateSimResults service does not produce a new samples file. In order to implement this behaviour with Argo, several workarounds need to be applied, which we describe in the following.
Argo is able to conditionally execute a workflow step. Since workflows are specified as so-called templates and a workflow step can be to execute a template, Argo allows each workflow template to call itself recursively. However, in contrast to our approach, the outputs of all workflow steps are hard-coded in the workflow template. This means that output files of the individual services are overwritten on each recursive call. One possibility to prevent this is to introduce a counter variable and to append its value to each output file name.
However, Argo has no support for counters and also does not offer a function to increment the value of a numerical variable. Similar to the file listing, the workflow designer needs to write an additional workflow step that consists of a Python program incrementing the counter and writing its new value to its standard output (see Fig. 24).
In addition, there is no possibility to stop the recursion based on the fact that a file does not exist (as necessary for the outer for-each action of this workflow). One possibility to work around this is to specify the name of the file (hard-coded with the counter value appended) as an output of the evaluateSimResults service and to specify a default value that will be applied if reading the file leads to an I/O error. This default value can then act as a break condition for the recursion. Note that this idea cannot exactly be mapped to how our system works because an I/O error does not necessarily mean that a file does not exist. It could be an actual fault (i.e. the network is temporarily unavailable), which would be ignored and would in fact lead to a successful workflow run. This can be dangerous as it hides actual errors and may lead to faulty simulation results.
Alternatively, the workflow designer can again write an additional workflow step with a Python program or a Bash script checking if the file exists and writing a value to its standard output.
Both approaches are workarounds that we seek to avoid with our solution. They make the workflow definition unnecessarily long and complex. Figure 25 compares the implementations of use case 2 in our system and Argo. Both systems use YAML. However, Argo’s definition file is not only longer in terms of the number of lines (three times as many lines) but some of them are also very long. This makes it hard for users to focus on the actual workflow. Also, the additional steps just needed to circumvent shortcomings of the system clutter the workflow definition and make it harder to maintain.
Summary
In “Goals and contributions” section, we defined two research questions. The first one was related to creating a distributed system that natively supports cyclic workflows without a priori run-time knowledge. The evaluation results above show that this research question can be successfully answered.
With the second research question, we wanted to investigate whether it is possible to design this system in a way so that cyclic workflows can be implemented without workarounds. We also wanted to show that this approach reduces design and maintenance effort. The implemented use cases above demonstrate that this is indeed possible. We summarised the differences between our system and the two tested ones in this regard in Table 6.
The table shows that our approach enables less complex workflow definitions since it offers native support for features that are important for the design and execution of cyclic workflows. For example, our system does not require the workflow designer to write additional generator scripts. It also does not need sub-workflows to simulate a dynamic workflow execution without a priori run-time knowledge.
Our system also does not require the workflow designer to keep an overview over hard-coded file names used within the workflow as well as over dependencies between actions. Our workflow model uses variables to store file names, and the system is able to automatically detect dependencies based on the usage of these variables.
In addition, our system is able to dynamically generate a list of file names from a directory during run time and to create a matching number of action instances to process these files in parallel. Pegasus and Argo require custom actions, sub-workflows/templates, or counter variables to simulate this behaviour.
To summarise, our system allows for writing workflow definitions that are much less complex and shorter than in the other systems. On the long run, these features help users in designing workflows and improves maintainability.
Conclusions
In this paper, we described an approach to scientific workflow management that supports complex, real-world scenarios where the structure of a workflow depends on the data to be processed and may change during run time. We presented an algorithm that traverses scientific workflow graphs and transforms them into independent linear sequences called process chains. We also presented a corresponding software architecture that executes the process chains in parallel in a distributed environment such as the cloud. Our system works iteratively and creates individual instances of workflow actions dynamically depending on both input data and data generated by preceding actions during run time. This enables support for the pattern “Multiple instances without a priori run-time knowledge” [13] and allows workflow designers to specify cycles in the workflow graph.
With the growing amount of global data, automated data processing solutions have become a fundamental tool. Data scientists often have to deal with a large number of heterogeneous data sets from various sources. In this respect, it is important that the complexity of the data processing pipeline stays at a maintainable level so the pipeline can be reused and extended over a long period of time. Compared to existing solutions, our approach does not rely on modelling directed acyclic graphs (DAGs) and does not require the workflow designer to implement special constructs for cycles and dynamically changing workflow structures. Instead, support for this is built into our solution, which leads to shorter, more reusable, and maintainable workflow designs. We have further shown that our approach allows for modelling generic workflows that can be applied to multiple input data sets without the need to change the static structure.
Our research questions defined in “Goals and contributions” section could therefore be answered successfully: It is possible to create a distributed scientific workflow management system that natively supports cyclic scientific workflows involving multiple instances of workflow actions without a priori run-time knowledge, and this native support leads to less complex workflows that do not require workarounds.
One of the building blocks of our approach is the lightweight workflow model presented in “Workflow model” section. This model consists of only two types of actions: execute and for-each. These actions are very generic and allow almost arbitrary workflows to be modelled. However, they are also limited in terms of expressiveness. For example, in our second use case in “Use case 2: shape optimisation via structural analysis” section, we used a for-each action to model multiple iterations that stop as soon as a certain condition becomes true, namely when the for-each action does not receive more input items from the last of its sub-actions. This break condition is implicit. In terms of maintainability, it would potentially be easier to comprehend if we had an explicit if action. Investigating the use of such an action remains for future work.
Another useful improvement to our system could be to unroll for-each actions only partially. This would help avoid having to keep all cloned instances of sub-actions in memory at the same time, which would further improve the scalability of our system, in particular if there is a large number of input items.
In this sense, we are continuously exploring new areas and improving our approach. The research results presented have evolved from a larger group of previous works: a microservice architecture for the processing of large geospatial data [3], workflow modelling with domain-specific languages [65], as well as capability-based task scheduling of process chains in a distributed environment [66]. As mentioned earlier, an implementation of our algorithm and our software architecture is available with the open-source workflow management system Steep [15]. The source code to reproduce our first use case is also available [58, 59].
Availability of data and materials
The source code of our algorithm and software architecture is available as open source on GitHub [15]. The code (and a script to download the 2MASS data) to reproduce the first use case from our evaluation is also available [58, 59]. We cannot provide data for the second use case as it is proprietary. However, the repository of Steep contains many test fixtures similar to this use case.
Notes
Personal communication with the corresponding author of [28], Marta Mattoso, 7 October 2020
Note that the condition also evaluates to true if the list is empty.
Abbreviations
- DAG:
-
Directed acyclic graph
- JVM:
-
Java virtual machine
- AWS:
-
Amazon web services
- 2MASS:
-
Two micron all sky survey
References
Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, Li P (2004) Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics 20(17):3045–3054. https://doi.org/10.1093/bioinformatics/bth361.
Graves R, Jordan TH, Callaghan S, Deelman E, Field E, Juve G, Kesselman C, Maechling P, Mehta G, Milner K, Okaya D, Small P, Vahi K (2011) Cybershake: A physics-based seismic hazard model for southern california. Pure Appl Geophys 168(3):367–381. https://doi.org/10.1007/s00024-010-0161-6.
Krämer M (2018) A microservice architecture for the processing of large geospatial data in the cloud. PhD thesis, Technische Universität Darmstadt. https://doi.org/10.13140/RG.2.2.30034.66248.
Krämer M, Senner I (2015) A modular software architecture for processing of big geospatial data in the cloud. Comput Graph 49:69–81. https://doi.org/10.1016/j.cag.2015.02.005.
Berriman GB, Deelman E, Good JC, Jacob JC, Katz DS, Kesselman C, Laity AC, Prince TA, Singh G, Su M-H (2004) Montage: a grid-enabled engine for delivering custom science-grade mosaics on demand In: Optimizing Scientific Return for Astronomy Through Information Technologies, vol. 5493, 221–233.. International Society for Optics and Photonics, Amsterdam.
Deelman E, Vahi K, Juve G, Rynge M, Callaghan S, Maechling PJ, Mayani R, Chen W, Ferreira da Silva R, Livny M, Wenger K (2015) Pegasus: a workflow management system for science automation. Futur Gener Comput Syst 46:17–35. https://doi.org/10.1016/j.future.2014.10.008.
Apache Airflow Documentation. https://airflow.apache.org/. Accessed 14 April 2020.
Zaharia M, Chowdhury M, Franklin MJ, Shenker S, Stoica I (2010) Spark: Cluster computing with working sets In: Proceedings of the 2nd USENIX Conference on Hot Topics in Cloud Computing, 1–10.
Carbone P, Katsifodimos A, Ewen S, Markl V, Haridi S, Tzoumas K (2015) Apache Flink: Stream and batch processing in a single engine. Bull IEEE Comput Soc Tech Comm Data Eng 36(4):28–38.
Rodriguez MA, Buyya R (2014) Deadline based resource provisioning and scheduling algorithm for scientific workflows on clouds. IEEE Trans Cloud Comput 2(2):222–235. https://doi.org/10.1109/TCC.2014.2314655.
Malawski M, Juve G, Deelman E, Nabrzyski J (2012) Cost- and deadline-constrained provisioning for scientific workflow ensembles in IaaS clouds In: Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, 1–11. https://doi.org/10.1109/SC.2012.38.
Bux M, Brandt J, Lipka C, Hakimzadeh K, Dowling J, Leser U (2015) SAASFEE: Scalable scientific workflow execution engine. Proc VLDB Endow 8(12):1892–1895. https://doi.org/10.14778/2824032.2824094.
Russell N, van van der Aalst WMP, ter Hofstede AHM (2016) Workflow Patterns: The Definitive Guide. MIT Press, Cambridge.
van der Aalst W, van Hee K (2004) Workflow Management: Models, Methods, and Systems. MIT Press, Cambridge.
Steep Workflow Management System. https://steep-wms.github.io/. Accessed 14 April 2020.
Deelman E, Peterka T, Altintas I, Carothers CD, van Dam KK, Moreland K, Parashar M, Ramakrishnan L, Taufer M, Vetter J (2018) The future of scientific workflows. Int J High Perform Comput Appl 32(1):159–175.
Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11(8):1–13.
Bernhardsson ELuigi Presentation NYC Data Science. https://www.slideshare.net/erikbern/luigi-presentation-nyc-data-science. Accessed 14 April 2020.
Balis B (2016) Hyperflow: A model of computation, programming approach and enactment engine for complex distributed workflows. Futur Gener Comput Syst 55:147–162.
Wozniak JM, Armstrong TG, Wilde M, Katz DS, Lusk E, Foster IT (2013) Swift/T: Large-scale application composition via distributed-memory dataflow processing In: 13th IEEE/ACM International Symposium on Cluster, Cloud, and Grid Computing, 95–102.. IEEE, New York City.
Zhao Y, Hategan M, Clifford B, Foster I, Von Laszewski G, Nefedova V, Raicu I, Stef-Praun T, Wilde M (2007) Swift: Fast, reliable, loosely coupled parallel computation In: IEEE Congress on Services (Services 2007), 199–206.. IEEE, New York City.
von Laszewski G, Hategan M (2005) Workflow concepts of the java CoG kit. J Grid Comput 3(3):239–258. https://doi.org/10.1007/s10723-005-9013-5.
Ogasawara E, Dias J, Silva V, Chirigati F, de Oliveira D, Porto F, Valduriez P, Mattoso M (2013) Chiron: a parallel engine for algebraic scientific workflows. Concurr Comput Pract Exp 25(16):2327–2341.
de Oliveira D, Ogasawara E, Baião F, Mattoso M (2010) Scicumulus: A lightweight cloud middleware to explore many task computing paradigm in scientific workflows In: 2010 IEEE 3rd International Conference on Cloud Computing, 378–385.. IEEE, New York City.
Ogasawara E, De Oliveira D, Valduriez P, Dias J, Porto F, Mattoso M (2011) An algebraic approach for data-centric scientific workflows. Proc VLDB Endowment (PVLDB) 4(11):1328–1339.
Souza R, Silva V, Coutinho AL, Valduriez P, Mattoso M (2017) Data reduction in scientific workflows using provenance monitoring and user steering. Futur Gener Comput Syst 110:481–501. https://doi.org/10.1016/j.future.2017.11.028.
Dias J, Ogasawara E, de Oliveira D, Porto F, Coutinho AL, Mattoso M (2011) Supporting dynamic parameter sweep in adaptive and user-steered workflow In: Proceedings of the 6th Workshop on Workflows in Support of Large-scale Science, 31–36.. Association for Computing Machinery, New York. https://doi.org/10.1145/2110497.2110502.
Dias J, Guerra G, Rochinha F, Coutinho AL, Valduriez P, Mattoso M (2015) Data-centric iteration in dynamic workflows. Futur Gener Comput Syst 46:114–126.
Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T (2006) Taverna: a tool for building and running workflows of services. Nucleic Acids Res 34:729–732.
Abouelhoda M, Issa SA, Ghanem M (2012) Tavaxy: Integrating taverna and galaxy workflows with cloud computing support. BMC Bioinformatics 13(1):1–19.
Altintas I, Berkley C, Jaeger E, Jones M, Ludascher B, Mock S (2004) Kepler: an extensible system for design and execution of scientific workflows In: 16th International Conference on Scientific and Statistical Database Management, 423–424.. IEEE, New York City.
Wang J, Altintas I (2012) Early cloud experiences with the kepler scientific workflow system. Proc Comput Sci 9:1630–1634.
Dean J, Ghemawat S (2008) MapReduce: simplified data processing on large clusters. Commun ACM 51(1):107–113.
Shvachko K, Kuang H, Radia S, Chansler R, et al (2010) The hadoop distributed file system In: MSST, vol. 10, 1–10.. IEEE, New York City. https://doi.org/10.1109/MSST.2010.5496972.
Wang J, Crawl D, Altintas I (2012) A framework for distributed data-parallel execution in the kepler scientific workflow system. Proc Comput Sci 9:1620–1629.
Fei X, Lu S, Lin C (2009) A mapreduce-enabled scientific workflow composition framework In: IEEE International Conference on Web Services, 663–670.. IEEE, New York City.
Souza R, Silva V, Miranda P, Lima A, Valduriez P, Mattoso M (2017) Spark scalability analysis in a scientific workflow In: Simpósio Brasileiro de Banco de Dados, 1–6.. UFC Brazil and UNI7 Brazil, Uberlandia, Minas Gerais.
Gaspar D, Porto F, Akbarinia R, Pacitti E (2017) Tardis: Optimal execution of scientific workflows in Apache Spark In: International Conference on Big Data Analytics and Knowledge Discovery, 74–87.. Springer, Cham.
van der Aalst WMP, ter Hofstede AHM (2005) YAWL: yet another workflow language. Inf Syst 30(4):245–275. https://doi.org/10.1016/j.is.2004.02.002.
Amstutz P, Crusoe MR, Tijanić N, Chapman B, Chilton J, Heuer M, Kartashov A, Kern J, Leehr D, Ménager H, Nedeljkovich M, Scales M, Soiland-Reyes S, Stojanovic L (2016) Common Workflow Language 1.0. https://doi.org/10.6084/m9.figshare.3115156.v2. Common Workflow Language Working Group.
Crusoe MRWPI’s Workflow Control Patterns and CWL. https://github.com/common-workflow-library/cwl-patterns/blob/794f96b/workflow_patterns_initiative/control/README.md . Accessed 6 Nov 2020.
Broad InstituteWorkflow Description Language 1.0. https://github.com/openwdl/wdl/blob/master/versions/1.0/SPEC.md. Accessed 14 April 2020.
Van der Aalst WMP (1998) The application of petri nets to workflow management. J Circ Syst Comput 8(1):21–66.
Adam NR, Atluri V, Huang W-K (1998) Modeling and analysis of workflows using petri nets. J Intell Inf Syst 10(2):131–158.
Salimifard K, Wright M (2001) Petri net-based modelling of workflow systems: An overview. Eur J Oper Res 134(3):664–676.
Giro S, Frydman C (2006) Modelling workflows using petri nets with multiple instances In: Proceedings of the Argentine Symposium on Computing Technology (AST).. UNLP, Mendoza.
The Vert.x ProjectVert.x. https://vertx.io/. Accessed 14 April 2020.
MongoDB IncMongoDB. https://www.mongodb.com/. Accessed 14 April 2020.
Red Hat Inc. https://www.gluster.org/. Accessed 14 April 2020.
HashiCorpTerraform. https://www.terraform.io/. Accessed 14 April 2020.
Red Hat IncAnsible – Simple IT Automation. https://www.ansible.com/. Accessed 14 April 2020.
Skrutskie MF, Cutri RM, Stiening R, Weinberg MD, Schneider S, Carpenter JM, Beichman C, Capps R, Chester T, Elias J, Huchra J, Liebert J, Lonsdale C, Monet DG, Price S, Seitzer P, Jarrett T, Kirkpatrick JD, Gizis JE, Howard E, Evans T, Fowler J, Fullmer L, Hurt R, Light R, Kopan EL, Marsh KA, McCallon HL, Tam R, Dyk SV, Wheelock S (2006) The two micron all sky survey (2MASS). Astron J 131(2):1163–1183. https://doi.org/10.1086/498708.
Montage Image Mosaic Engine. http://montage.ipac.caltech.edu/. Accessed 14 April 2020.
Deelman E, Singh G, Livny M, Berriman B, Good J (2008) The cost of doing science on the cloud: The Montage example In: Proceedings of the ACM/IEEE Conference on Supercomputing, 1–12. https://doi.org/10.1109/SC.2008.5217932.
Tanaka M, Tatebe O (2012) Workflow scheduling to minimize data movement using multi-constraint graph partitioning In: 12th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing, 65–72. https://doi.org/10.1109/CCGrid.2012.134.
Llwaah F, Thomas N, Cała J (2015) Improving MCT scheduling algorithm to reduce the makespan and cost of workflow execution in the cloud In: 31st UK Performance Engineering Workshop.. University of Leeds, UK.
Bharathi S, Chervenak A, Deelman E, Mehta G, Su M, Vahi K (2008) Characterization of scientific workflows In: Third Workshop on Workflows in Support of Large-Scale Science, 1–10. https://doi.org/10.1109/WORKS.2008.4723958.
Montage Workflow with Steep. https://github.com/steep-wms/steep-montage. Accessed 15 April 2020.
AWS Configuration Files for the Montage Workflow with Steep. https://github.com/steep-wms/steep-montage-aws. Accessed 15 April 2020.
Tamellini L, Chiumenti M, Altenhofen C, Attene M, Barrowclough O, Livesu M, Marini F, Martinelli M, Skytt V (2019) Parametric shape optimization for combined additive–subtractive manufacturing. JOM. https://doi.org/10.1007/s11837-019-03886-x.
Joy KI, MacCracken R (1999) The Refinement Rules for Catmull-Clark Solids. Technical Report CSE-96-1, Department of Computer Science, University of California.
Altenhofen C, Schuwirth F, Stork A, Fellner D (2017) Volumetric subdivision for consistent implicit mesh generation. Comput Graph 69:68–79. https://doi.org/10.1016/j.cag.2017.09.005.
Altenhofen C, Loosmann F, Mueller-Roemer JS, Grasser T, Luu TH, Stork A (2017) Integrating interactive design and simulation for mass customized 3d-printed objects – a cup holder example In: Solid Freeform Fabrication Symposium, vol. 28, 2289–2301.. University of Texas, Austin.
Montage Workflow with Pegasus. https://github.com/pegasus-isi/montage-workflow-v2. Accessed 10 Aug 2020.
Krämer M (2014) Controlling the processing of smart city data in the cloud with domain-specific languages In: Proceedings of the 7th International Conference on Utility and Cloud Computing UCC, 824–829.. IEEE. https://doi.org/10.1109/UCC.2014.134.
Krämer M (2020) Capability-based scheduling of scientific workflows in the cloud In: Proceedings of the 9th International Conference on Data Science, Technology, and Applications DATA, 43–54.. SciTePress. https://doi.org/10.5220/0009805400430054.
Acknowledgements
This publication makes use of data products from the Two Micron All Sky Survey, which is a joint project of the University of Massachusetts and the Infrared Processing and Analysis Center/California Institute of Technology, funded by the National Aeronautics and Space Administration and the National Science Foundation.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Authors’ contributions
Michel Krämer is the main author of the manuscript. He has designed the algorithm and the software architecture and implemented them in the open-source workflow management system Steep. Hendrik M. Würz has contributed “Related work” and “Comparison with Pegasus” sections and helped Michel Krämer with fruitful discussions during the algorithm design phase. Christian Altenhofen has implemented the shape optimisation workflow to evaluate the approach presented in this paper. He wrote “Use case 2: shape optimisation via structural analysis” section summarising his findings. The author(s) read and approved the final manuscript.
Authors’ information
Michel Krämer
Dr. Michel Krämer is deputy head of the competence centre for Spatial Information Management at the Fraunhofer Institute for Computer Graphics Research IGD. He received a PhD in Computer Science from the Technical University of Darmstadt. His research interests are in processing of big geospatial data, microservices, Cloud Computing, and DevOps. Michel Krämer has contributed to a number of EC-funded research projects. Michel Krämer is development lead of several open source projects and contributes to popular frameworks and libraries.
Hendrik M. Würz
Hendrik M. Würz is a researcher at the Fraunhofer Institute for Computer Graphics Research IGD and studies visual computing at the Technical University of Darmstadt. He is interested in the distributed processing of big data and the possibilities of Function as a Service. Hendrik M. Würz has experience in the preparation of geodata for visualisation and the usage of cloud-based resources.
Christian Altenhofen
Christian Altenhofen, M.Sc. is a senior researcher at the Fraunhofer Institute for Computer Graphics Research IGD and PhD student at Technical University of Darmstadt. As part of the competence centre for “Interactive Engineering Technologies” at IGD, he works on improving product development and manufacturing processes by developing novel methods in the areas of Computer-Aided Design (CAD), Computer-Aided Engineering (CAE), and Isogeometric Analysis (IGA). In the past seven years, Christian Altenhofen has participated in several EC-funded, as well as nationally and regionally funded research projects, two of which focussed on bringing engineering services to the Cloud.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krämer, M., Würz, H.M. & Altenhofen, C. Executing cyclic scientific workflows in the cloud. J Cloud Comp 10, 25 (2021). https://doi.org/10.1186/s13677-021-00229-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13677-021-00229-7



 AND-split
AND-split AND-join
AND-join OR-split
OR-split AND/OR-split
AND/OR-split For-each action
For-each action




















